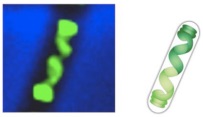
Bacteria are arguably the simplest living organisms that are able to reproduce independently. Most bacteria possess one circular chromosome that is 1000 times compacted occupying only about 15% of cell volume. This pseudo-compartment of chromosome is known as nucleoid and is maintained in absence of any nuclear membrane, unlike eukaryotes. Surprisingly, we know more about cells of higher organisms (eukaryotic) than we do in bacteria (prokaryotic). How the E. coli DNA, e.g., that comprises of 4.6 Mbp and has a length of about 1.5mm in the extended state fits into a cell of linear dimension of about is not well understood. The nucleoid was earlier perceived to be an amorphous entity composed of random positioning of ill-defined domains of supercoils. Recent experiments, however, showed a remarkable degree of spatial organization [e.g, Berlatzky et. al. PNAS 105, 14136 (2008), where from the figure on the left is adapted]
We have recently shown that entropic forces can drive such spatial organization of chromosome [Phys Rev Lett, (2012)]. We considered a bottle-brush arrangement of DNA with a main-chain attached with side loops to model cross-linking in DNA due to chromosome modification proteins. We showed that such a model with an open linear backbone spontaneously adopts a helical structure with a well-defined pitch when confined to small cylindrical volume of the bacterial cell. The results were analyzed in terms of the interplay between the effective stiffness and actual intra-chain packing effects caused by the side loops in response to the confinement. We envisage that this structural arrangement plays a crucial role in the segregation dynamics of the newly replicated bacterial chromosome.